Among the most intensively studied loci to date is the locus encoding the cystic fibrosis transmembrane conductance regulator (CFTR). Recessive loss-of-function mutations in the CFTR locus cause cystic fibrosis, a disease with symptoms including salty skin and the production of an excess of thick mucus in the lungs, which leads to susceptibility to bacterial infections. Geneticists have examined the CFTR locus in some 30,000 chromosomes from individuals with cystic fibrosis and have found more than 500 different mutations that can cause the disease. Among these mutations are missense mutations, amino acid deletions, nonsense mutations, frameshifts, and splice defects.
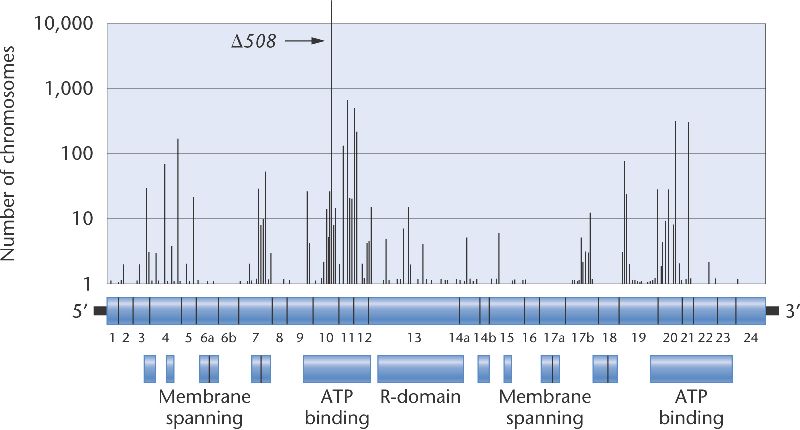
Figure 26-3 shows a map of the 27 exons in the CFTR locus, with most exons identified by function. The histogram above the map shows the locations of some of the disease causing mutations and the number of copies of each that have been found. A single mutation, a 3-bp deletion in exon 10 called δ508 accounts for 67 percent of all mutant cystic fibrosis alleles, but several other mutations are found in at least 100 of the chromosomes surveyed. In populations of European ancestry, between 1 in 44 and 1 in 20 individuals are heterozygous carriers of mutant alleles. Note that Figure 26-3 includes only the sequence variants that alter the function of the CFTR protein. There are undoubtedly many more CFTR alleles with silent sequence variants that do not change the structure of the protein and that do not affect its function.
Figure 26-3. The locations of disease-causing mutations in the cystic fibrosis gene. The histogram shows the number of copies of each mutation geneticists have found. (The vertical axis is on a logarithmic scale.) The genetic map below the histogram shows the locations and relative sizes of the 27 exons of the CFTR locus. The boxes at the bottom indicate the functions of different domains of the CFTR protein.