[Page 658] Researchers investigating this question have used information derived from nucleotide sequencing of the mitochondrial genes for small-subunit ribosomal RNAs (SSU rRNA). These sequences are under strong functional constraint and so are highly conserved. Phylogenetic analysis of the sequences identified a group of bacteria known as the α-proteobacteria (purple bacteria) as the closest living relatives of mitochondria. More recent work has divided this group into two subclasses, one of which is called the rickettsial subdivision.
Based on the SSU phylogeny, the rickettsia have been identified as the bacteria most closely related to mitochondria. Interestingly, rickettsia, like mitochondria, live only inside eukaryotic cells.
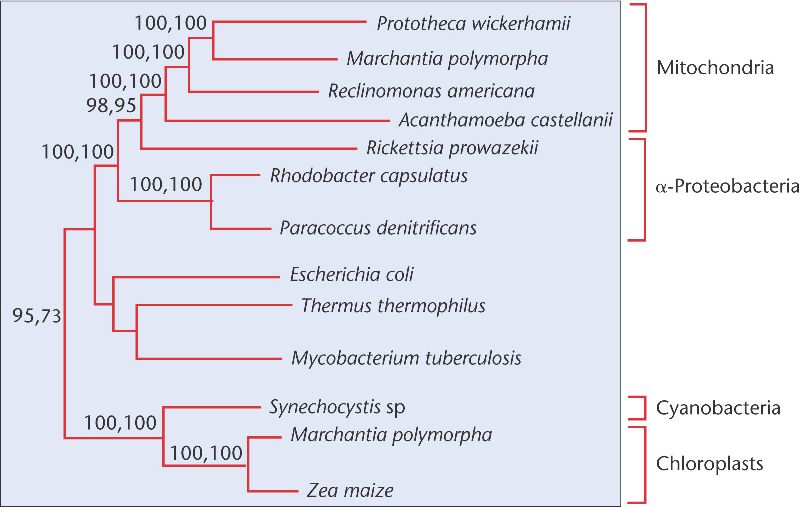
Unlike mitochondria, however, rickettsia are disease-causing parasites, responsible for typhus, one of the most serious diseases in the history of our species. Genomic sequencing of one rickettsial species, Rickettsia prowazekii, provides additional insight into the relationship between these bacteria and mitochondria. Using the genome sequence information for genes involved in ATP synthesis derived from R. prowazekii, other bacteria, and mitochondria from several sources, Siv Andersson and colleagues constructed a phylogenetic tree (Figure 26-23). Their tree indicates a close evolutionary relationship between R. prowazekii and mitochondria. Evidence from SSU analysis results in a similar tree. Thus, two lines of evidence, from SSUs and proteins involved in ATP synthesis, provide strong evidence that the mitochondria we carry in our cells derive from an ancient ancestor of the rickettsia.
Figure 26-23. The evolutionary relationships of mitochondria, α-proteobacteria, cyanobacteria, and chloroplasts. Note that, although R. prowazekii is an α-proteobacterium it is more closely related to mitochondria than to other α-proteobacteria.