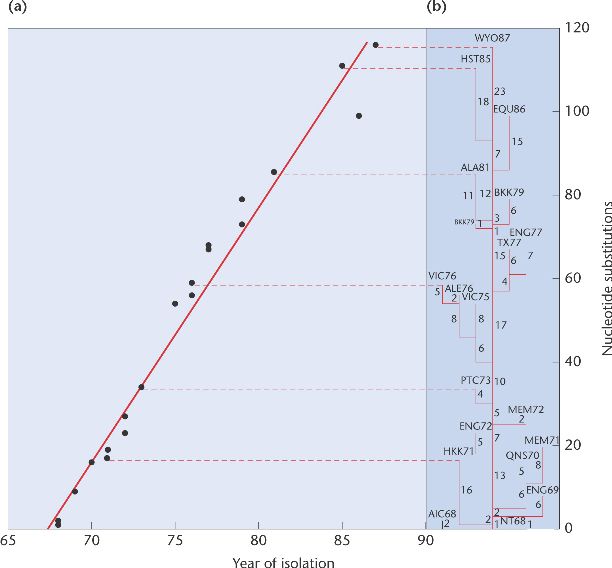
Research by Walter M. Fitch and colleagues on the influenza A virus shows how molecular clocks are used. Fitch and his associates sequenced part of the hemagglutinin gene from flu viruses that had been isolated at different times over a 20-year period. They calculated the number of nucleotide differences among the various viruses and constructed an evolutionary tree [Figure 26-20(b)]. Most strains have gone extinct, leaving no descendants among the more recently isolated viruses. Fitch's group then plotted the number of nucleotide substitutions between the first virus and each subsequent virus against the year in which the virus was isolated [Figure 26-20(a)]. The points all fall very close to a straight line, indicating that nucleotide substitutions in this gene have accumulated at a steady rate. The hemagglutinin gene thus serves as a molecular clock. Molecular clocks are used to compare the sequences of new flu viruses as they appear each year and to estimate the time that has passed since each diverged from a common ancestor.
[Page 656] Figure 26-20. Molecular clock in the influenza A hemagglutinin gene. (a) Number of nucleotide differences between the first isolate and each subsequent isolate as a function of year of isolation. (b) Estimate of the phylogeny of the isolates.